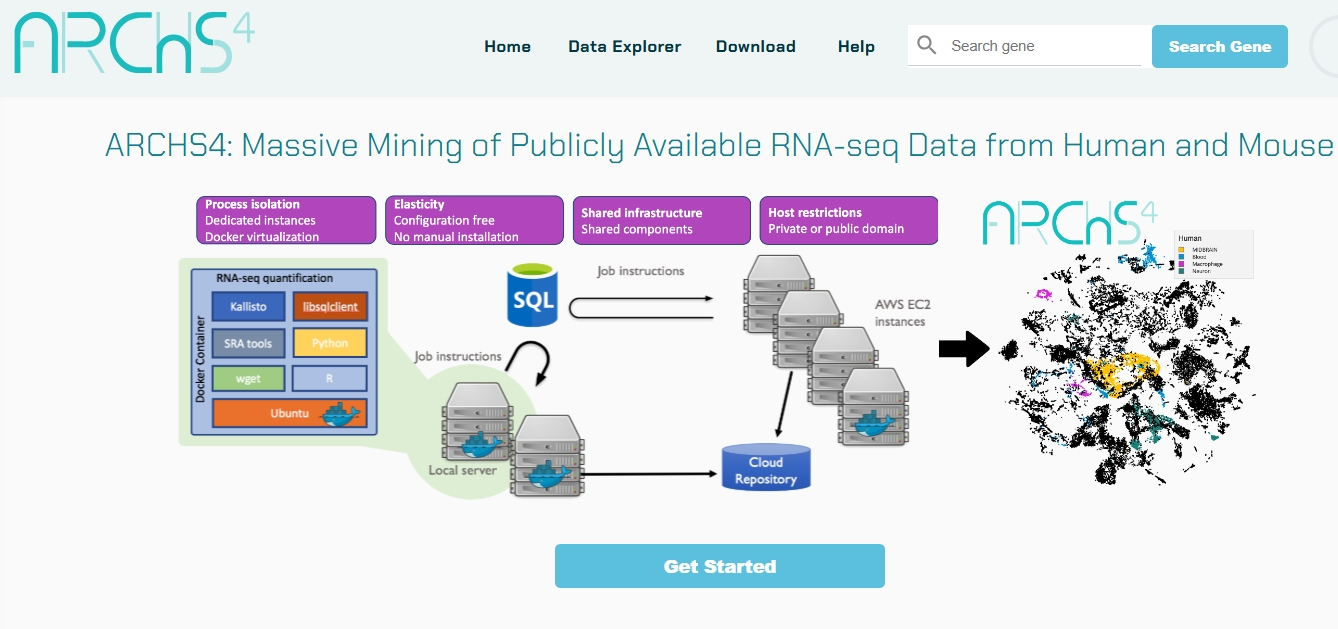
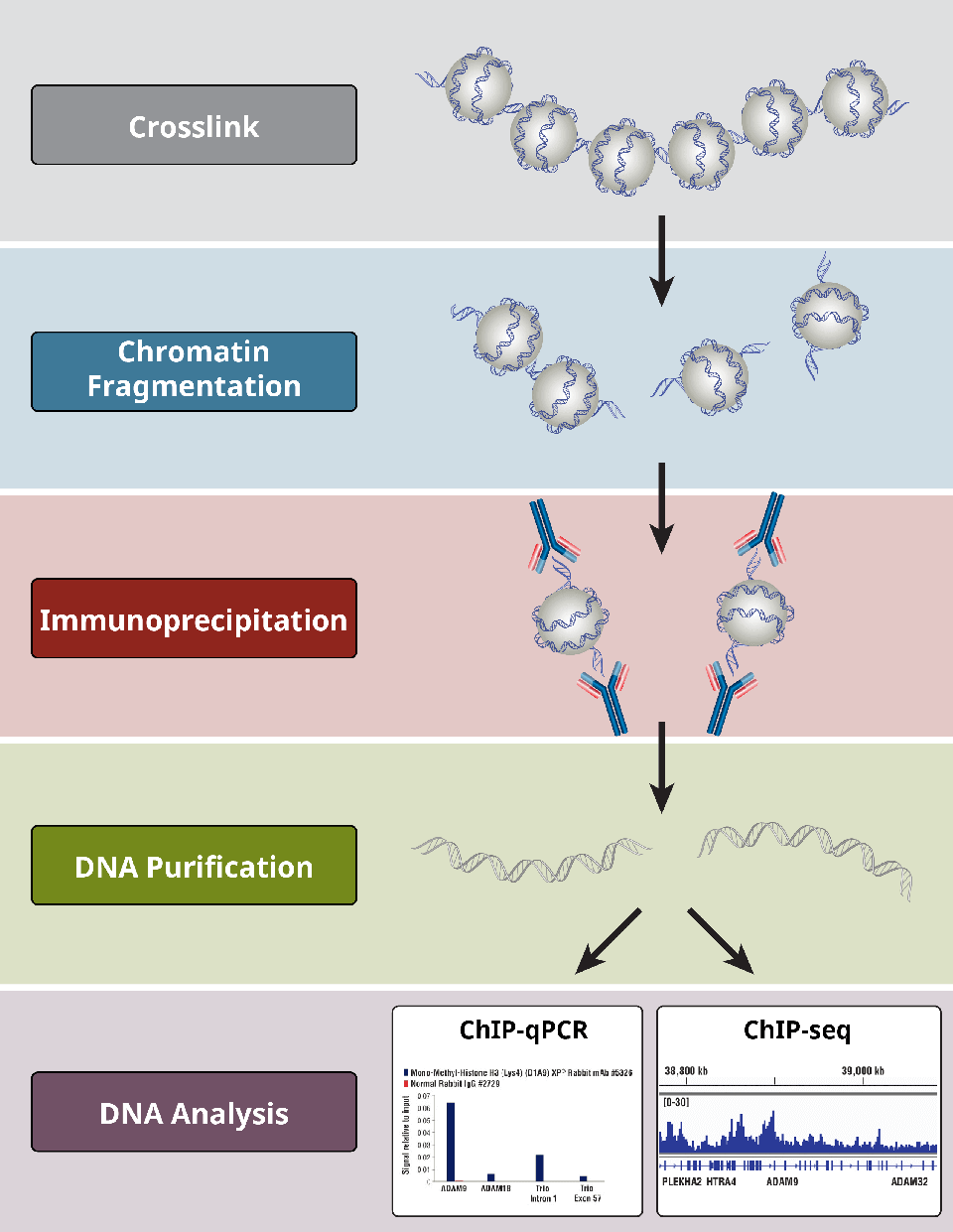
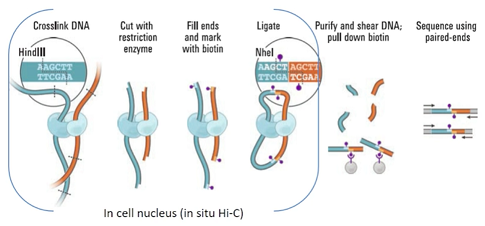
error in .rs.downloadfile(url = url, destfile = path, method = method)
新装的系统,刚开始装包都挺顺利了,今天突然遇到
error in .rs.downloadfile(url = url, destfile = path, method = method),不管用devtools::install_github还是remotes方法,都不能安装github上的包。
查了好多办法没解决,也看了很多配置都是正确的。
后来尝试把method改成git,就可以下载了,这也说明系统底层网络是通的,问题出在方法上。默认的方式是libcurl。libcurl需要SSL/TLS 协议。问题锁定在 R 内部对 libcurl 的调用配置上。R在用libcurl的时候,会调用系统的证书库进行,但现在R 不知道去哪里找系统的证书库。当它尝试连接 GitHub 的 HTTPS 地址时,无法验证 GitHub 的 SSL 证书,出于安全机制会直接报错中断。
|
|
既然有证书,那就让系统知道证书的位置
|
|
这样就可以解决了。但始终不清楚为什么系统找不到证书库这个问题,费解。