Python抓取动态网页
生物信息学中,David(the Database for Annotation,Visualization and Integrated Discovery)是常用的注视工具,可以对基因进行GO注释,KEGG pathway注释等,David提供接口供批量注释调用。
David的网址https://david.ncifcrf.gov/,api介绍https://david.ncifcrf.gov/content.jsp?file=DAVID_API.html
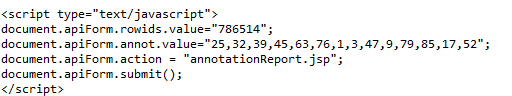
不是生物信息学的朋友,可以重点关注分析思路。 以Entrez gene id为1002的基因为例,返回GO,interpro等注释信息的api格式为
用python应用urllib2的包,抓取上述网页的结果为