MAF格式Mutation Annotation Format (MAF) ,是TCGA组织对突变进行注释的格式。一些和癌症分析相关的软件,经常要求MAF格式文件作为输入。而现在经过GATK或samtools检测出突变的格式一般为VCF格式,的注释软件,即使经过SNPEff和annovar注释(当然还有VEP),结果依然为VCF格式或者tab分割的文件等。
MAF中每一列是一种注释信息,由于包含的注释信息太多(详见格式),单纯的通过写脚本转换SNPEff或者annovar的注释文件,会变得非常麻烦而且考虑的问题可能不完全(有人实现过,通过Ensembl的VEP对VCF注释,然后转换,可以在github上搜索到)。
这里介绍注释软件oncotator,可以注释VCF文件,并直接生成MAF格式,相当于将VCF格式转换成MAF格式。Broad institute开发的,用起来放心哈。
首先安装python virtualenv
virtualenv可以创建一个虚拟的python环境,在虚拟环境中安装的包,不影响机器已经安装的python包。
1
2
3
4
5
6
|
curl -O https://pypi.python.org/packages/source/v/virtualenv/virtualenv-14.0.6.tar.gz
tar -xvzf virtualenv-14.0.6.tar.gz
cd virtualenv-14.0.6/
python setup.py install --prefix=~/pypackages/
cd ..
rm -rf virtualenv-14.0.6*
|
安装oncotator依赖
1
2
3
4
|
git clone https://github.com/broadinstitute/oncotator.git
cd oncotator
mkdir env
bash scripts/create_oncotator_venv.sh -e ./env
|
安装oncotator
1
2
3
|
source ./env/bin/activate
python setup.py install
deactivate
|
下载注释数据文件
1
2
|
wget -c http://www.broadinstitute.org/~lichtens/oncobeta/oncotator_v1_ds_Jan262015.tar.gz
tar -xvzf oncotator_v1_ds_Jan262015.tar.gz
|
注释内容如下,包括基因组注释,蛋白注释,癌症突变注释等,用的基因组是GENCODE的hg19版本。
Genomic Annotations
Gene, transcript, and functional consequence annotations using GENCODE for hg19.
Reference sequence around a variant.
GC content around a variant.
Human DNA Repair Gene annotations from Wood et al.
Protein Annotations
Site-specific protein annotations from UniProt.
Functional impact predictions from dbNSFP.
Cancer Variant Annotations
Observed cancer mutation frequency annotations from COSMIC.
Cancer gene and mutation annotations from the Cancer GenCensus.
Overlapping mutations from the Cancer Cell Line Encyclopedia.
Cancer gene annotations from the Familial Cancer Database.
Cancer variant annotations from ClinVar.
Non-Cancer Variant Annotations
Common SNP annotations from dbSNP.
Variant annotations from 1000 Genomes.
Variant annotations from NHLBI GO Exome Sequencing Project (ESP).
运行oncotator
1
2
3
4
5
6
|
#进入虚拟环境env
source ./env/bin/activate
# WGS = whole genome sequencing, WXS = whole exome sequencing
# 通过annotate-manual设置MAF注释,比如样本barcode,样本来源等,至于氨基酸突变等,会自动进行注释
oncotator --input_format=VCF --db-dir=./oncotator_v1_ds_Jan262014 --output_format=TCGAMAF --tx-mode=EFFECT --collapse-number-annotations --annotate-default='Sequencer:Illumina HiSeq' --annotate-manual='Sequence_Source:WGS' --annotate-manual='Tumor_Sample_Barcode:Tumor' --annotate-manual='Matched_Norm_Sample_Barcode:normal' input.vcf output.maf hg19
deactivate
|
结果
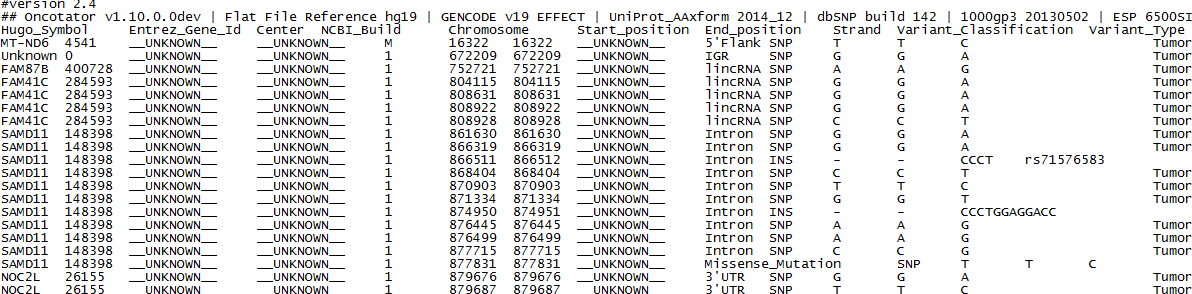
下面的内容如果在浏览器中有换行,可以直接复制到文本编辑器中看。
1
2
3
4
5
6
7
8
9
10
|
#version 2.4
## Oncotator v1.10.0.0dev ' Flat File Reference hg19 ' GENCODE v19 EFFECT ' UniProt_AAxform 2014_12 ' dbSNP build 142 ' 1000gp3 20130502 ' ESP 6500SI-V2 ' ESP 6500SI-V2 ' dbNSFP v2.4 ' COSMIC v62_291112 ' UniProt_AA 2014_12 ' ORegAnno UCSC Track ' CCLE_By_GP 09292010 ' ClinVar 12.03.20 ' Ensembl ICGC MUCOPA ' ACHILLES_Lineage_Results 110303 ' COSMIC_Tissue 291112 ' CGC full_2012-03-15 ' TCGAScape 110405 ' HGNC Sept172014 ' Familial_Cancer_Genes 20110905 ' CCLE_By_Gene 09292010 ' MutSig Published Results 20110905 ' UniProt 2014_12 ' COSMIC_FusionGenes v62_291112 ' HumanDNARepairGenes 20110905 ' TUMORScape 20100104 ' gencode_xref_refseq metadata_v19
Hugo_Symbol Entrez_Gene_Id Center NCBI_Build Chromosome Start_position End_position Strand Variant_Classification Variant_Type Reference_Allele Tumor_Seq_Allele1 Tumor_Seq_Allele2 dbSNP_RS dbSNP_Val_Status Tumor_Sample_Barcode Matched_Norm_Sample_Barcode Match_Norm_Seq_Allele1 Match_Norm_Seq_Allele2 Tumor_Validation_Allele1 Tumor_Validation_Allele2 Match_Norm_Validation_Allele1 Match_Norm_Validation_Allele2 Verification_Status Validation_Status Mutation_Status Sequencing_Phase Sequence_Source Validation_Method Score BAM_file Sequencer Tumor_Sample_UUID Matched_Norm_Sample_UUID Genome_Change Annotation_Transcript Transcript_Strand Transcript_Exon Transcript_Position cDNA_Change Codon_Change Protein_Change Other_Transcripts Refseq_mRNA_Id Refseq_prot_Id SwissProt_acc_Id SwissProt_entry_Id Description UniProt_AApos UniProt_Region UniProt_Site UniProt_Natural_Variations UniProt_Experimental_Info GO_Biological_Process GO_Cellular_Component GO_Molecular_Function COSMIC_overlapping_mutations COSMIC_fusion_genes COSMIC_tissue_types_affected COSMIC_total_alterations_in_gene Tumorscape_Amplification_Peaks Tumorscape_Deletion_Peaks TCGAscape_Amplification_Peaks TCGAscape_Deletion_Peaks DrugBank ref_context gc_contentCCLE_ONCOMAP_overlapping_mutations CCLE_ONCOMAP_total_mutations_in_gene CGC_Mutation_Type CGC_Translocation_Partner CGC_Tumor_Types_Somatic CGC_Tumor_Types_Germline CGC_Other_Diseases DNARepairGenes_Role FamilialCancerDatabase_Syndromes MUTSIG_Published_Results OREGANNO_ID OREGANNO_Values 1000gp3_AA 1000gp3_AC 1000gp3_AF 1000gp3_AFR_AF 1000gp3_AMR_AF 1000gp3_AN 1000gp3_CIEND 1000gp3_CIPOS 1000gp3_CS 1000gp3_DP 1000gp3_EAS_AF 1000gp3_EN1000gp3_EUR_AF 1000gp3_IMPRECISE 1000gp3_MC 1000gp3_MEINFO 1000gp3_MEND 1000gp3_MLEN 1000gp3_MSTART 1000gp3_NS1000gp3_SAS_AF 1000gp3_SVLEN 1000gp3_SVTYPE 1000gp3_TSD ACHILLES_Lineage_Results_Top_Genes CGC_Cancer Germline Mut CGC_Cancer Molecular Genetics CGC_Cancer Somatic Mut CGC_Cancer Syndrome CGC_Chr CGC_Chr Band CGC_GeneID CGC_Name CGC_Other Germline Mut CGC_Tissue Type COSMIC_n_overlapping_mutations COSMIC_overlapping_mutation_descriptions COSMIC_overlapping_primary_sites ClinVar_ASSEMBLY ClinVar_HGMD_ID ClinVar_SYM ClinVar_TYPE ClinVar_rs ESP_AA ESP_AAC ESP_AA_AC ESP_AA_AGE ESP_AA_GTC ESP_AvgAAsampleReadDepth ESP_AvgEAsampleReadDepth ESP_AvgSampleReadDepth ESP_CA ESP_CDP ESP_CG ESP_CP ESP_Chromosome ESP_DBSNP ESP_DP ESP_EA_AC ESP_EA_AGE ESP_EA_GTC ESP_EXOME_CHIP ESP_FG ESP_GL ESP_GM ESP_GS ESP_GTC ESP_GTS ESP_GWAS_PUBMED ESP_MAF ESP_PH ESP_PP ESP_Position ESP_TAC ESP_TotalAAsamplesCovered ESP_TotalEAsamplesCovered ESP_TotalSamplesCovered Ensembl_so_accession Ensembl_so_term Familial_Cancer_Genes_Reference Familial_Cancer_Genes_Synonym HGNC_Accession Numbers HGNC_CCDS IDs HGNC_Chromosome HGNC_Date Modified HGNC_Date Name Changed HGNC_Date Symbol Changed HGNC_Ensembl Gene ID HGNC_Ensembl ID(supplied by Ensembl) HGNC_Enzyme IDs HGNC_Gene family description HGNC_HGNC ID HGNC_Locus Group HGNC_Locus Type HGNC_Name Synonyms HGNC_OMIM ID(supplied by NCBI) HGNC_Previous Names HGNC_Previous Symbols HGNC_Primary IDs HGNC_Pubmed IDs HGNC_Record Type HGNC_RefSeq(supplied by NCBI) HGNC_Secondary IDs HGNC_Status HGNC_Synonyms HGNC_UCSC ID(supplied by UCSC) HGNC_UniProt ID(supplied by UniProt) HGNC_VEGA IDs HGVS_coding_DNA_change HGVS_genomic_change HGVS_protein_change ORegAnno_bin UniProt_alt_uniprot_accessions alt_allele_seen annotation_transcript build ccds_id dbNSFP_1000Gp1_AC dbNSFP_1000Gp1_AF dbNSFP_1000Gp1_AFR_AC dbNSFP_1000Gp1_AFR_AF dbNSFP_1000Gp1_AMR_AC dbNSFP_1000Gp1_AMR_AF dbNSFP_1000Gp1_ASN_AC dbNSFP_1000Gp1_ASN_AF dbNSFP_1000Gp1_EUR_AC dbNSFP_1000Gp1_EUR_AF dbNSFP_Ancestral_allele dbNSFP_CADD_phred dbNSFP_CADD_raw dbNSFP_CADD_raw_rankscore dbNSFP_ESP6500_AA_AF dbNSFP_ESP6500_EA_AF dbNSFP_Ensembl_geneid dbNSFP_Ensembl_transcriptid dbNSFP_FATHMM_pred dbNSFP_FATHMM_rankscore dbNSFP_FATHMM_score dbNSFP_GERP++_NR dbNSFP_GERP++_RS dbNSFP_GERP++_RS_rankscore dbNSFP_Interpro_domain dbNSFP_LRT_Omega dbNSFP_LRT_converted_rankscore dbNSFP_LRT_pred dbNSFP_LRT_score dbNSFP_LR_pred dbNSFP_LR_rankscore dbNSFP_LR_score dbNSFP_MutationAssessor_pred dbNSFP_MutationAssessor_rankscore dbNSFP_MutationAssessor_score dbNSFP_MutationTaster_converted_rankscore dbNSFP_MutationTaster_pred dbNSFP_MutationTaster_score dbNSFP_Polyphen2_HDIV_pred dbNSFP_Polyphen2_HDIV_rankscore dbNSFP_Polyphen2_HDIV_score dbNSFP_Polyphen2_HVAR_pred dbNSFP_Polyphen2_HVAR_rankscore dbNSFP_Polyphen2_HVAR_score dbNSFP_RadialSVM_pred dbNSFP_RadialSVM_rankscoredbNSFP_RadialSVM_score dbNSFP_Reliability_index dbNSFP_SIFT_converted_rankscore dbNSFP_SIFT_pred dbNSFP_SIFT_score dbNSFP_SLR_test_statistic dbNSFP_SiPhy_29way_logOdds dbNSFP_SiPhy_29way_logOdds_rankscore dbNSFP_SiPhy_29way_pi dbNSFP_UniSNP_ids dbNSFP_Uniprot_aapos dbNSFP_Uniprot_acc dbNSFP_Uniprot_id dbNSFP_aaalt dbNSFP_aapos dbNSFP_aapos_FATHMM dbNSFP_aapos_SIFT dbNSFP_aaref dbNSFP_cds_strand dbNSFP_codonpos dbNSFP_fold-degenerate dbNSFP_genename dbNSFP_hg18_pos(1-coor) dbNSFP_phastCons100way_vertebrate dbNSFP_phastCons100way_vertebrate_rankscore dbNSFP_phastCons46way_placental dbNSFP_phastCons46way_placental_rankscore dbNSFP_phastCons46way_primate dbNSFP_phastCons46way_primate_rankscore dbNSFP_phyloP100way_vertebrate dbNSFP_phyloP100way_vertebrate_rankscore dbNSFP_phyloP46way_placental dbNSFP_phyloP46way_placental_rankscore dbNSFP_phyloP46way_primate dbNSFP_phyloP46way_primate_rankscore dbNSFP_refcodon entrez_gene_id gc_content_full gencode_transcript_name gencode_transcript_status gencode_transcript_tags gencode_transcript_type gene_type havana_transcript id qual refseq_mrna_id secondary_variant_classification
MT-ND6 4541 __UNKNOWN__ __UNKNOWN__ M 16322 16322 __UNKNOWN__ 5'Flank SNP T T C Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chrM:16322T>C ENST00000361681.2 - 0 0 MT-TP_ENST00000387461.2_RNA'MT-TE_ENST00000387459.1_RNA'MT-TT_ENST00000387460.2_RNA P03923 NU6M_HUMAN mitochondrially encoded NADH dehydrogenase 6 cellular metabolic process (GO:0044237)'respiratory electron transport chain (GO:0022904)'small molecule metabolic process (GO:0044281) integral component of membrane (GO:0016021)'mitochondrial inner membrane (GO:0005743)'respiratory chain (GO:0070469) NADH dehydrogenase (ubiquinone) activity (GO:0008137) breast(1)'endometrium(10)'kidney(17)'urinary_tract(1) 29 GTACATAAAGTCATTTACCGT 0.478 SO:0001631 upstream_gene_variant mitochondria 2011-07-04 2005-02-15 2005-02-16ENSG00000198695 ENSG00000198695 1.6.5.3 """Mitochondrial respiratory chain complex / Complex I""" 7462 protein-coding gengene with protein product """complex I ND6 subunit"", ""NADH-ubiquinone oxidoreductase chain 6""" 516006 """NADH dehydrogenase 6""" MTND6 Standard Approved NAD6, ND6 P03923 chrM.hg19:g.16322T>C Exception_encountered Q34774'Q8HG30 True ENST00000361681.2 hg19 0.478 MT-ND6-201 KNOWN basic'appris_principal protein_coding protein_coding 185.84 YP_003024037
Unknown 0 __UNKNOWN__ __UNKNOWN__ 1 672209 672209 __UNKNOWN__ IGR SNP G G A Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:672209G>A RP5-857K21.4 (16629 upstream) : RP11-206L10.3 (4983 downstream) taaggacagcgcaaccttgat 0.493 SO:0001628 intergenic_variant chr1.hg19:g.672209G>A True hg19 0.493 rs79279787 56.77
FAM87B 400728 __UNKNOWN__ __UNKNOWN__ 1 752721 752721 __UNKNOWN__ lincRNA SNP A A G Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:752721A>G ENST00000326734.1 + 0 0 RP11-206L10.10_ENST00000435300.1_RNA NR_103536.1 family with sequence similarity 87, member B AGAAAGGAAAAGGGAAGAATG 0.488 .''' 3272 0.653355 0.2905 0.7363 5008 , , 22729 0.7659 0.839 False ,,2504 0.7781 0 AK097327 1p36.33 2013-02-01 ENSG00000177757 ENSG00000177757 """Long non-coding RNAs""" 32236 non-coding RNA RNA, long non-coding Standard NR_103536 Approved FLJ40008 Q8N852 OTTHUMG00000002471 chr1.hg19:g.752721A>G True ENST00000326734.1 hg19 0.488 FAM87B-001 KNOWN basic lincRNA lincRNA OTTHUMT00000007025.1 rs3131972 951.77
FAM41C 284593 __UNKNOWN__ __UNKNOWN__ 1 804115 804115 __UNKNOWN__ lincRNA SNP G G A Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:804115G>A ENST00000446136.1 - 0 1202 NR_027055.family with sequence similarity 41, member C AATCCCTAGAGACAACCTACC 0.368 .''' 4587 0.915935 0.8699 0.9236 5008 , , 17607 0.8423 0.993 False ,,2504 0.9693 0 BC047940 1p36.33 2013-01-16 2011-08-31 2011-08-31 ENSG00000230368 ENSG00000230368 """Long non-coding RNAs""" 27635 non-coding RNA RNA, long non-coding 12477932 Standard NR_027055 Approved uc001abt.4 OTTHUMG00000002469 chr1.hg19:g.804115G>A True ENST00000446136.1 hg19 0.368 FAM41C-001 KNOWN basic lincRNA lincRNA OTTHUMT00000007021.1 rs9725068 68.77 NR_027055
FAM41C 284593 __UNKNOWN__ __UNKNOWN__ 1 808631 808631 __UNKNOWN__ lincRNA SNP G G A Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:808631G>A ENST00000446136.1 - 0 1202 NR_027055.family with sequence similarity 41, member C ACTGATGACCGTATACCTGGC 0.547 .''' 2271 0.453474 0.115 0.6527 5008 , , 21503 0.4167 0.7724 False ,,2504 0.4796 0 BC047940 1p36.33 2013-01-16 2011-08-31 2011-08-31 ENSG00000230368 ENSG00000230368 """Long non-coding RNAs""" 27635 non-coding RNA RNA, long non-coding 12477932 Standard NR_027055 Approved uc001abt.4 OTTHUMG00000002469 chr1.hg19:g.808631G>A True ENST00000446136.1 hg19 0.547 FAM41C-001 KNOWN basic lincRNA lincRNA OTTHUMT00000007021.1 rs11240779 412.77 NR_027055
FAM41C 284593 __UNKNOWN__ __UNKNOWN__ 1 808922 808922 __UNKNOWN__ lincRNA SNP G G A Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:808922G>A ENST00000446136.1 - 0 1202 NR_027055.family with sequence similarity 41, member C CCAAGGTGTCGGACACCGTGG 0.532 .''' 4921 0.982628 0.9554 0.9942 5008 , , 21602 0.9841 1.0 False ,,2504 0.9918 0 BC047940 1p36.33 2013-01-16 2011-08-31 2011-08-31 ENSG00000230368 ENSG00000230368 """Long non-coding RNAs""" 27635 non-coding RNA RNA, long non-coding 12477932 Standard NR_027055 Approved uc001abt.4 OTTHUMG00000002469 chr1.hg19:g.808922G>A True ENST00000446136.1 hg19 0.532 FAM41C-001 KNOWN basic lincRNA lincRNA OTTHUMT00000007021.1 rs6594027 815.77 NR_027055
FAM41C 284593 __UNKNOWN__ __UNKNOWN__ 1 808928 808928 __UNKNOWN__ lincRNA SNP C C T Tumore normal __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ WGS __UNKNOWN__ __UNKNOWN__ __UNKNOWN__ Illumina HiSeq Tumore N63_S1.variant g.chr1:808928C>T ENST00000446136.1 - 0 1202 NR_027055.family with sequence similarity 41, member C TGTCGGACACCGTGGTGGAGC 0.517 .''' 2265 0.452276 0.1172 0.6527 5008 , , 22141 0.4157 0.7734 False ,,2504 0.4703 0 BC047940 1p36.33 2013-01-16 2011-08-31 2011-08-31 ENSG00000230368 ENSG00000230368 """Long non-coding RNAs""" 27635 non-coding RNA RNA, long non-coding 12477932 Standard NR_027055 Approved uc001abt.4 OTTHUMG00000002469 chr1.hg19:g.808928C>T True ENST00000446136.1 hg19 0.517 FAM41C-001 KNOWN basic lincRNA lincRNA OTTHUMT00000007021.1 rs11240780 259.77 NR_027055
|
参考
http://dx.doi.org/10.1002/humu.22771
http://portals.broadinstitute.org/oncotator/
这个网页里面包含一个自动安装依赖的脚本
http://gatkforums.broadinstitute.org/gatk/discussion/4153/what-are-the-platform-requirements-and-software-dependencies-for-running-oncotator
If you want to create indexed VCF files for use with Oncotator, you will need to have Samtools and Tabix installed and on your path.
http://gatkforums.broadinstitute.org/gatk/discussion/4154/howto-install-and-run-oncotator-for-the-first-time
http://gatkforums.broadinstitute.org/gatk/discussion/4162/howto-set-up-a-virtual-environment-and-install-all-dependencies-for-oncotator
https://wiki.nci.nih.gov/display/TCGA/Mutation+Annotation+Format+%28MAF%29+Specification;WIKISESSIONID=52395E56D6223DF1AB5840164882D34A
https://github.com/broadinstitute/oncotator
####################################################################
#版权所有 转载请告知 版权归作者所有 如有侵权 一经发现 必将追究其法律责任
#Author: Jason
###################################################################

