miRNA biogenesis and function
文章目录
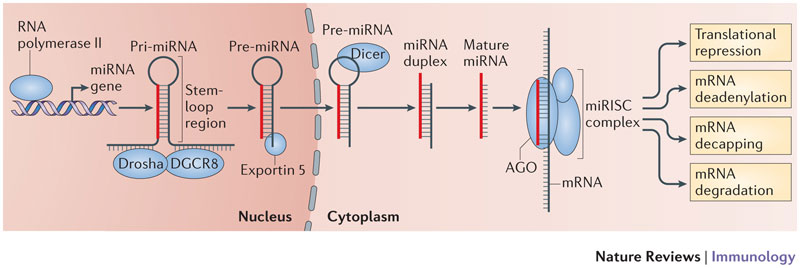
MicroRNA (miRNA) genes are transcribed into primary miRNAs (pri-miRNAs) by RNA polymerase II. Pri-miRNAs are bound by the microprocessor complex subunit DGCR8 and processed by the ribonuclease III (RNase III) activity of Drosha into hairpin structures known as precursor miRNAs (pre-miRNAs). Exportin 5 shuttles pre-miRNAs from the nucleus into the cytoplasm where the RNase III Dicer cleaves off the hairpin loop of the pre-miRNA. The resulting duplex segregates and the mature single-stranded miRNA associates with Argonaute (AGO) proteins and other accessory proteins to form the miRNA-induced silencing complex (miRISC), which mediates the translational repression and the increased degradation of its mRNA targets. A mature miRNA bound to an AGO protein forms the core of the miRISC. The AGO protein recruits other protein complexes that antagonize translation and that deadenylate the targeted mRNA130. This ultimately leads to mRNA decapping and degradation. Therefore, the effect of miRNA-mediated repression can be observed at both the protein and the mRNA level (see figure).
The miRNA provides specificity through complementary base pairing with target mRNAs11. Nucleotides in positions 2-8 from the 5ʹ end of an miRNA (termed the seed sequence) are a major determinant of target recognition. However, complementarity in the 3ʹ half of the miRNA can contribute to binding and ‘seedless’ targets that rely on non-seed sequences for binding also occur. Most functional miRNA-binding sites occur in the 3ʹ untranslated region of target mRNAs and many of these are highly conserved, which indicates that miRNAs have co-evolved with their targets. These principles have been exploited to develop algorithms for the bioinformatic prediction of miRNA targets. Although these algorithms are useful for hypothesis generation, they remain imperfect; any predicted targets must be experimentally confirmed and many true targets are missed.
文章作者 zzx
上次更新 2013-09-06
