I always use gtf file and retrieve gene information. There isn’t a highly flexible tool to solve my demand. I modified the code from “https://github.com/Jverma/GFF-Parser”, thanks Jverma. This tool will be easier to use.
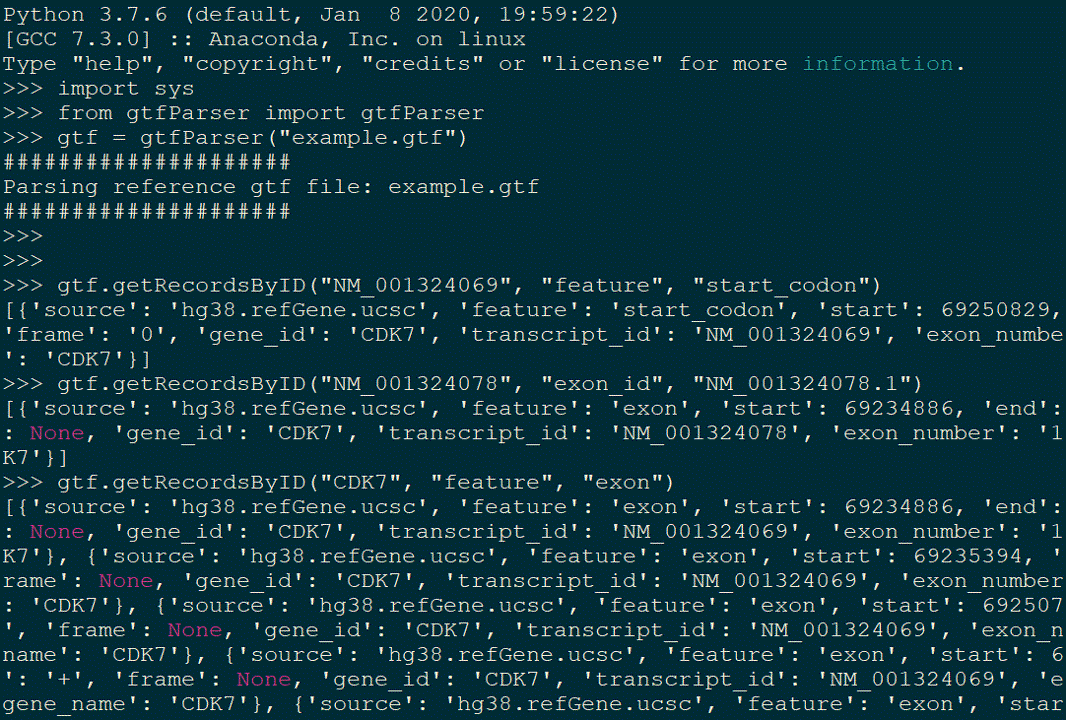
Usage
Basically, there are three parameters.
id: either transcript id or gene id.
attType: attribute defined in gtf file. E.g. feature (column 3), gene_name, exon_number, transcript_id in column 9
attValue: the attribute value you want to search for.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
>>> import sys
>>> from gtfParser import gtfParser
>>> gtf = gtfParser("example.gtf")
>>> # Get all exons in CDK7
>>> gtf.getRecordsByID("CDK7", "feature", "exon")
>>> # Get all features of transcript_id defined as "NM_001324069" in gene "CDK7"
>>> gtf.getRecordsByID("CDK7", "transcript_id", "NM_001324069")
>>> # Get start codon where feature was defined as "start_codon" in transcript "NM_001324069"
>>> gtf.getRecordsByID("NM_001324069", "feature", "start_codon")
>>> # Get a exon where its id is "NM_001324078.1" in "NM_001324078" transcript
>>> gtf.getRecordsByID("NM_001324078", "exon_id", "NM_001324078.1")
|
Example gtf
Here is an simple example of gtf file. You can use to test. A subset from refSeq.hg38.gtf.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
|
chr5 hg38.refGene.ucsc transcript 69234886 69277430 . + . gene_id "CDK7"; transcript_id "NM_001324069"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69234886 69235041 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "1"; exon_id "NM_001324069.1"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69235394 69235453 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "2"; exon_id "NM_001324069.2"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69250745 69250831 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "3"; exon_id "NM_001324069.3"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69250829 69250831 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "3"; exon_id "NM_001324069.3"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69252418 69252451 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "4"; exon_id "NM_001324069.4"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69252418 69252451 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "4"; exon_id "NM_001324069.4"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69254602 69254669 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "5"; exon_id "NM_001324069.5"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69254602 69254669 . + 2 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "5"; exon_id "NM_001324069.5"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69255460 69255528 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "6"; exon_id "NM_001324069.6"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69255460 69255528 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "6"; exon_id "NM_001324069.6"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69258043 69258153 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "7"; exon_id "NM_001324069.7"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69258043 69258153 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "7"; exon_id "NM_001324069.7"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69259818 69259936 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "8"; exon_id "NM_001324069.8"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69259818 69259936 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "8"; exon_id "NM_001324069.8"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69262205 69262304 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "9"; exon_id "NM_001324069.9"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69262205 69262304 . + 1 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "9"; exon_id "NM_001324069.9"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69269207 69269293 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "10"; exon_id "NM_001324069.10"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69269207 69269293 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "10"; exon_id "NM_001324069.10"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69272892 69273041 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "11"; exon_id "NM_001324069.11"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69272892 69273041 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "11"; exon_id "NM_001324069.11"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69276543 69276690 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "12"; exon_id "NM_001324069.12"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69276543 69276690 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "12"; exon_id "NM_001324069.12"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69277107 69277430 . + . gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "13"; exon_id "NM_001324069.13"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69277107 69277132 . + 2 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "13"; exon_id "NM_001324069.13"; gene_name "CDK7";
chr5 hg38.refGene.ucsc start_codon 69250829 69250831 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "3"; exon_id "NM_001324069.3"; gene_name "CDK7";
chr5 hg38.refGene.ucsc stop_codon 69277133 69277135 . + 0 gene_id "CDK7"; transcript_id "NM_001324069"; exon_number "13"; exon_id "NM_001324069.13"; gene_name "CDK7";
chr5 hg38.refGene.ucsc transcript 69234886 69277430 . + . gene_id "CDK7"; transcript_id "NM_001324078"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69234886 69235041 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "1"; exon_id "NM_001324078.1"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69235394 69235453 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "2"; exon_id "NM_001324078.2"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69254602 69254667 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "3"; exon_id "NM_001324078.3"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69255460 69255528 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "4"; exon_id "NM_001324078.4"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69255511 69255528 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "4"; exon_id "NM_001324078.4"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69258043 69258153 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "5"; exon_id "NM_001324078.5"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69258043 69258153 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "5"; exon_id "NM_001324078.5"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69259818 69259936 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "6"; exon_id "NM_001324078.6"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69259818 69259936 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "6"; exon_id "NM_001324078.6"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69262205 69262304 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "7"; exon_id "NM_001324078.7"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69262205 69262304 . + 1 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "7"; exon_id "NM_001324078.7"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69269207 69269293 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "8"; exon_id "NM_001324078.8"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69269207 69269293 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "8"; exon_id "NM_001324078.8"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69272892 69273041 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "9"; exon_id "NM_001324078.9"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69272892 69273041 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "9"; exon_id "NM_001324078.9"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69276543 69276690 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "10"; exon_id "NM_001324078.10"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69276543 69276690 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "10"; exon_id "NM_001324078.10"; gene_name "CDK7";
chr5 hg38.refGene.ucsc exon 69277107 69277430 . + . gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "11"; exon_id "NM_001324078.11"; gene_name "CDK7";
chr5 hg38.refGene.ucsc CDS 69277107 69277132 . + 2 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "11"; exon_id "NM_001324078.11"; gene_name "CDK7";
chr5 hg38.refGene.ucsc start_codon 69255511 69255513 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "4"; exon_id "NM_001324078.4"; gene_name "CDK7";
chr5 hg38.refGene.ucsc stop_codon 69277133 69277135 . + 0 gene_id "CDK7"; transcript_id "NM_001324078"; exon_number "11"; exon_id "NM_001324078.11"; gene_name "CDK7";
|
My code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
|
#!/usr/bin/env python
# -*- coding: utf-8 -*-
#############################################################
### Inspired by https://github.com/Jverma/GFF-Parser ###
#############################################################
####################################################################
# >>> from gtfParser import gtfParser
# >>> gtf = gtfParser(input_file)
#
# >>> gtf.getRecordsByID("TTN", "feature", "exon")
# >>> gtf.getRecordsByID("TTN", "transcript_id", "NM_001367552")
# >>> gtf.getRecordsByID("NM_013416", "feature", "start_codon")
# >>> gtf.getRecordsByID("NR_024540", "exon_id", "NR_024540.5")
####################################################################
class gtfParser:
def __init__(self, input_file):
import sys
self.data = {}
self.dict = {}
self.gene_attributes_dict = {} # ZZX
self.transcriptID_geneID_dict = {} # ZZX
sys.stderr.write("#####################\nParsing reference gtf file: " +
input_file +
'\n#####################\n')
for line in open(input_file):
if line.startswith("#"): continue
record = line.strip().split("\t")
sequence_name = record[0]
source = record[1]
feature = record[2]
start = int(record[3])
end = int(record[4])
if (record[5] != '.'):
score = record[5]
else:
score = None
strand = record[6]
if (record[7] != '.'):
frame = record[7]
else:
frame = None
attributes = record[8].split(';') # please note the separator between each attribute
attributes = [x.strip() for x in attributes[0:-1]] # ZZX
attributes = {x.split(' ')[0]: x.split(' ')[1].strip("\"") for x in attributes if " " in x} # ZZX
if not (sequence_name in self.data): self.data[sequence_name] = []
alpha = {'source': source, 'feature': feature, 'start': start, 'end': end, 'score': score, 'strand': strand,
'frame': frame}
# python 3 .items(), python 2 .iteritems() ZZX
for k, v in attributes.items(): alpha[k] = v
self.data[sequence_name].append(alpha)
# ZZX
for k, v in self.data.items():
for alpha in v:
gene_id = alpha["gene_id"]
transcript_id = alpha["transcript_id"]
self.transcriptID_geneID_dict[transcript_id] = gene_id
if gene_id in self.gene_attributes_dict.keys():
self.gene_attributes_dict[gene_id].append(alpha)
else:
self.gene_attributes_dict[gene_id] = list()
self.gene_attributes_dict[gene_id].append(alpha)
def getRecordsByID(self, id, attType, attValue):
"""
Gets all the features for a given gene
:param id: Identifier of the gene (specified by gene_id) or mRNA (specified by transcript_id)
:param attType: Any attribute list in gtf file.
:param attValue:
:return: A list of features related to ID where restricted by attType and attValue.
"""
att_list = []
if id in self.gene_attributes_dict.keys():
for x in self.gene_attributes_dict[id]:
if ( attType in x.keys() and x[attType] == attValue ):
att_info = x
att_list.append(att_info)
elif id in self.transcriptID_geneID_dict.keys():
for x in self.gene_attributes_dict[self.transcriptID_geneID_dict[id]]:
if ( attType in x.keys() and x[attType] == attValue and x["transcript_id"] == id):
att_info = x
att_list.append(att_info)
else:
sys.stderr.write("Could not find ID "+id+'\n')
sys.exit(1)
return att_list
|
Code available in GitHub:
https://github.com/ProfessionalFarmer/lake/blob/master/gtfParser.py
#####################################################################
#版权所有 转载请告知 版权归作者所有 如有侵权 一经发现 必将追究其法律责任
#Author: Jason
#####################################################################

