Prepare a data frame for sample CNV data
文章目录
If we want to cluster samples based on CNV data, a dataframe is needed. However, CNV segments in each sample are not the same. Maybe overlap or distinct. I think CNTools package migh solve this challenge. An example is shown as below. The result is a reduced segment data frame.
|
|
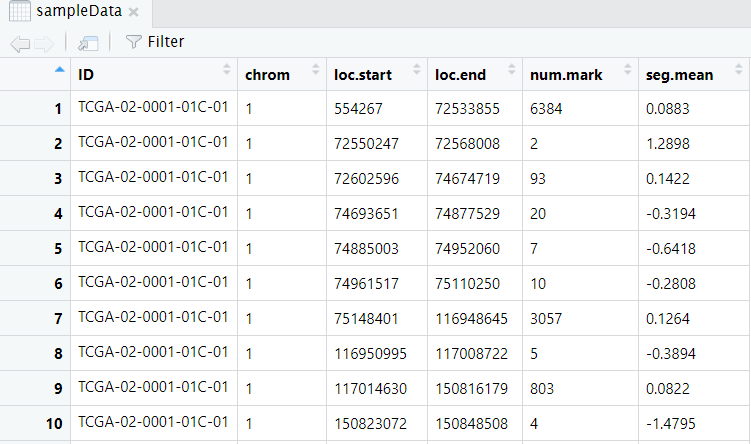
Input dataframe has six columns (“ID”,“chrom”,“loc.start”,“loc.end”,“num.mark”,“seg.mean”) including 277 samples and 54825 segments.
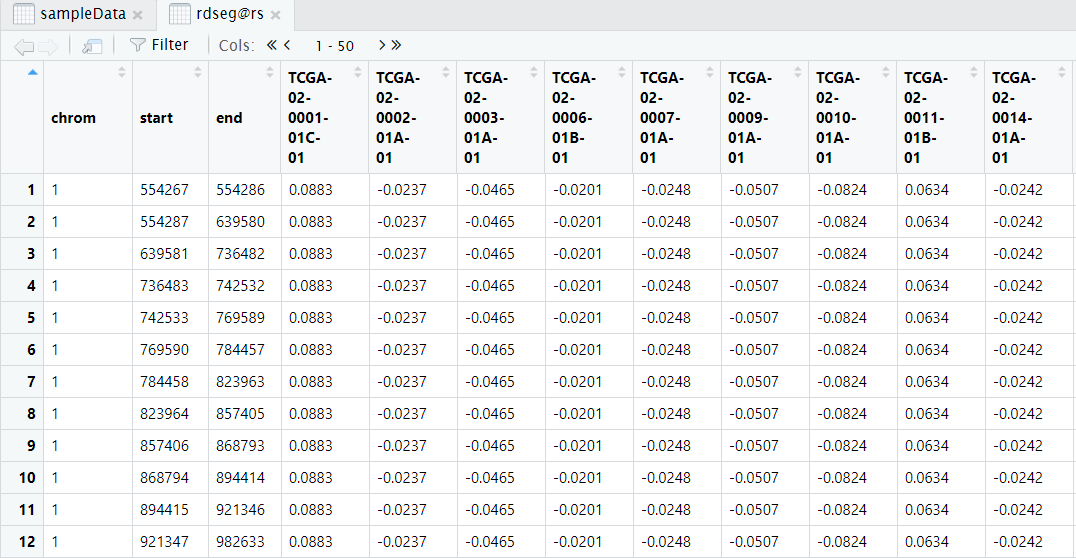
The result can be got from rdseg@rs, like this
 Cheers Also, we can use CNRegions from iClusterPlus package. CNregions(sampleData)
Cheers Also, we can use CNRegions from iClusterPlus package. CNregions(sampleData)
Ref: https://www.rdocumentation.org/packages/CNTools https://rdrr.io/bioc/iClusterPlus/man/CNregions.html ##################################################################### #版权所有 转载请告知 版权归作者所有 如有侵权 一经发现 必将追究其法律责 #Author: Jason
################################################################
文章作者 zzx
上次更新 2020-03-26
